AggreCount
An automated image analysis tool

An unbiased automated image analysis tool for quantification and localization of cellular aggregates
AggreCount is an automated image analysis tool written in the ImageJ macro language based on the FIJI distribution of ImageJ (version 1.52p or later). AggreCount requires dual channel images that have at least immunohistochemical stains for aggregates and nuclei though it supports n-channel images.
Advanced image processing
AggreCount utilizes the difference of gaussians (DoG) methodology to isolate puncta from immunohistochemical images. DoG allows for the reliable quantification of puncta even when images have uneven background illumination, out of focus fields, or varying fluorescence intensities. AC is ready for analysis out of the box for a large variety of aggregate stains (inclduing ubiquitin stains) though may be modified by users to analyze numerous other punctate structures such as peroxisomes and lysosomes.
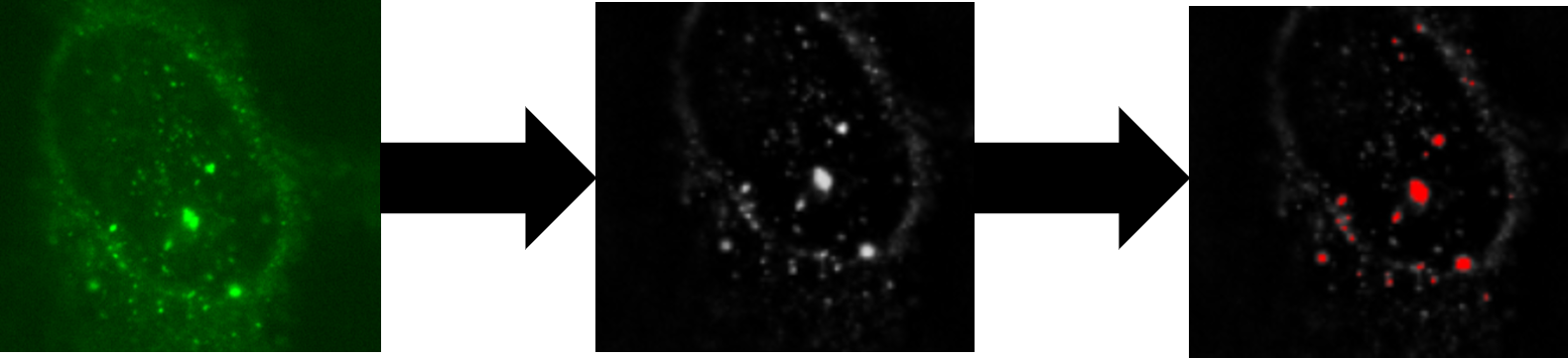
Accelerated aggregate quantification
AggreCount is ready for batch image analysis allowing for rapid quantification of 100s of images. AC can accept any file type supported by the BioFormats image importer including proprietary image file types such as .nd2. Find all supported formats in the BioFormats documentation
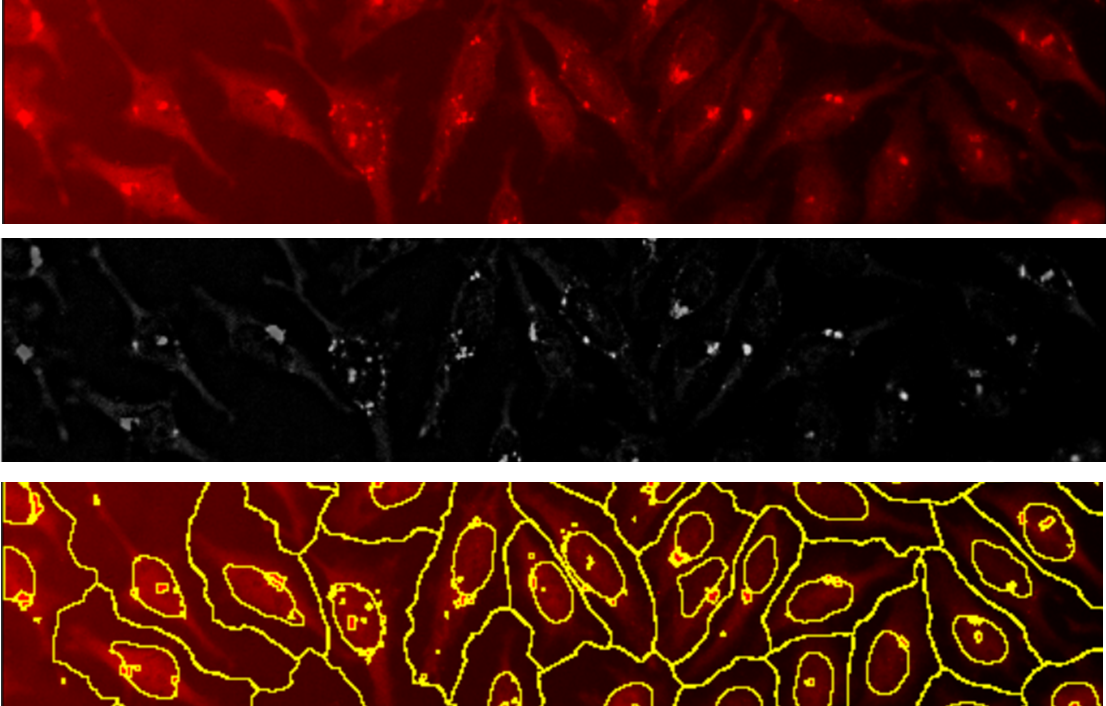
Easy cell segmentation
AggreCount uses the watershed-based segmentation algorithm native to FIJI allowing for segmentation of cells without the need of machine learning algorithms and training sets. This segmentation methods allows for separation of cells even when closely packed. Users may change the ‘prominence’ variable that determines local maxima which defines cells using the main settings window.
AggreCount on BioRx
The AggreCount journal article is now in preprint on BioRx.
Developed in the Raman Lab at Tufts University
AggreCount was developed by Jacob Klickstein in the Raman Lab in 2020.
This work was supported by NIH grant R01GM127557 to Malavika Raman Ph.D.
Find more information on the lab, our research, and our members by visiting the lab website here Raman Lab
